6.7 EMP_marker_analysis
The module EMP_marker_analysis is designed to utilize machine learning algorithms to identify signature features within multi-omics data. It is important to note that in omics data analysis, machine learning models primarily serve two purposes: the first is to use machine learning algorithms to filter out features with potential marker capabilities; the second is to construct machine learning predictive models based on omics data, to assist in distinguishing between different groups. Since building predictive models often requires multiple parameter tuning optimizations, using default parameters can often lead to unsatisfactory results. Therefore, the design of module EMP_marker_analysis aims to assist users in quickly screening potential markers, rather than focusing on building and optimizing predictive models.
6.7.1 Estimate the importance of features based on the Boruta algorithm
🏷️Example:
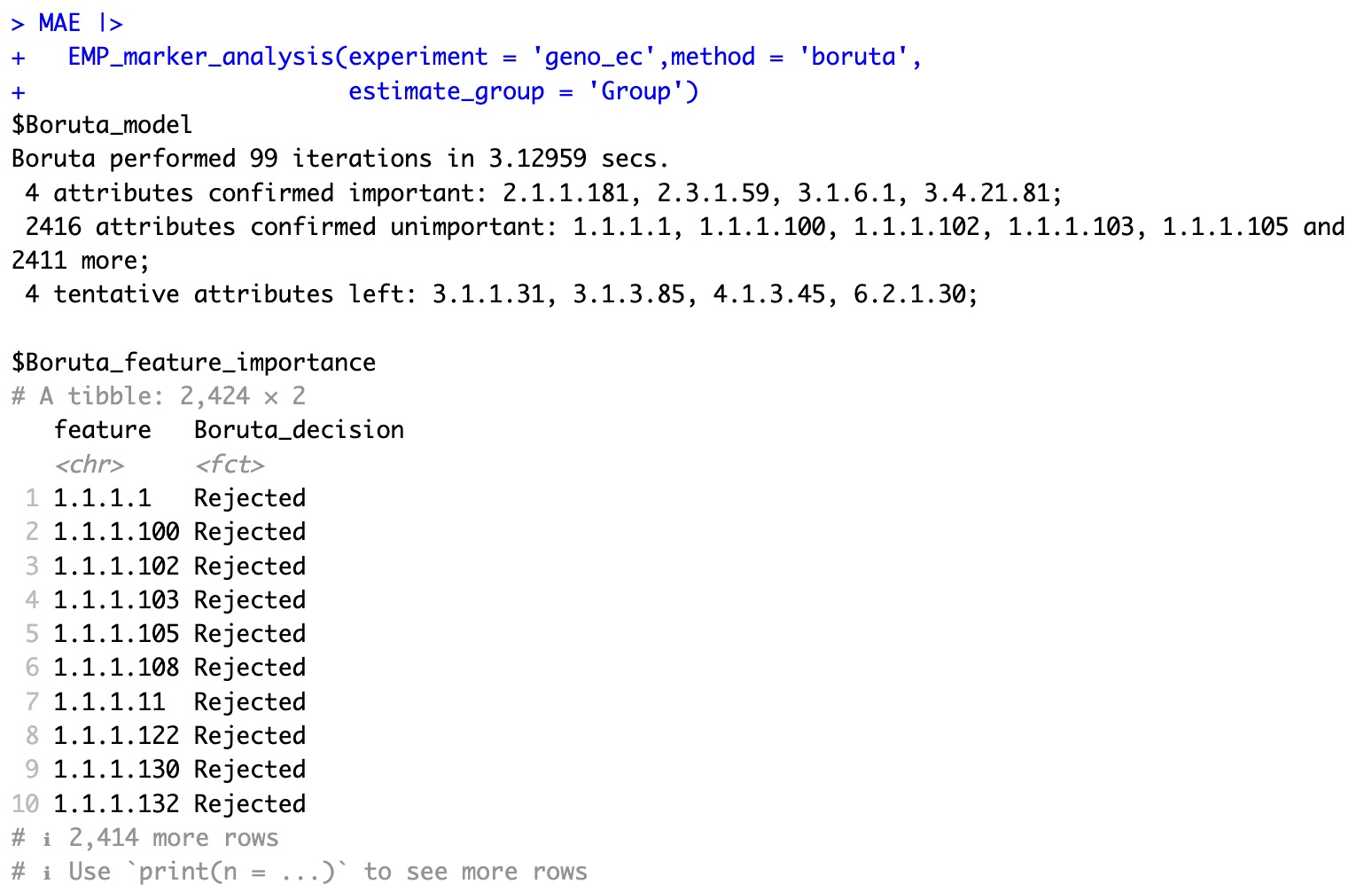
The Boruta algorithm can be used to quickly categorize the features in the data into three categories, Confirmed , Tentative , and Rejected , based on the classification in coldata.
MAE |>
EMP_marker_analysis(experiment = 'geno_ec',method = 'boruta',
estimate_group = 'Group')
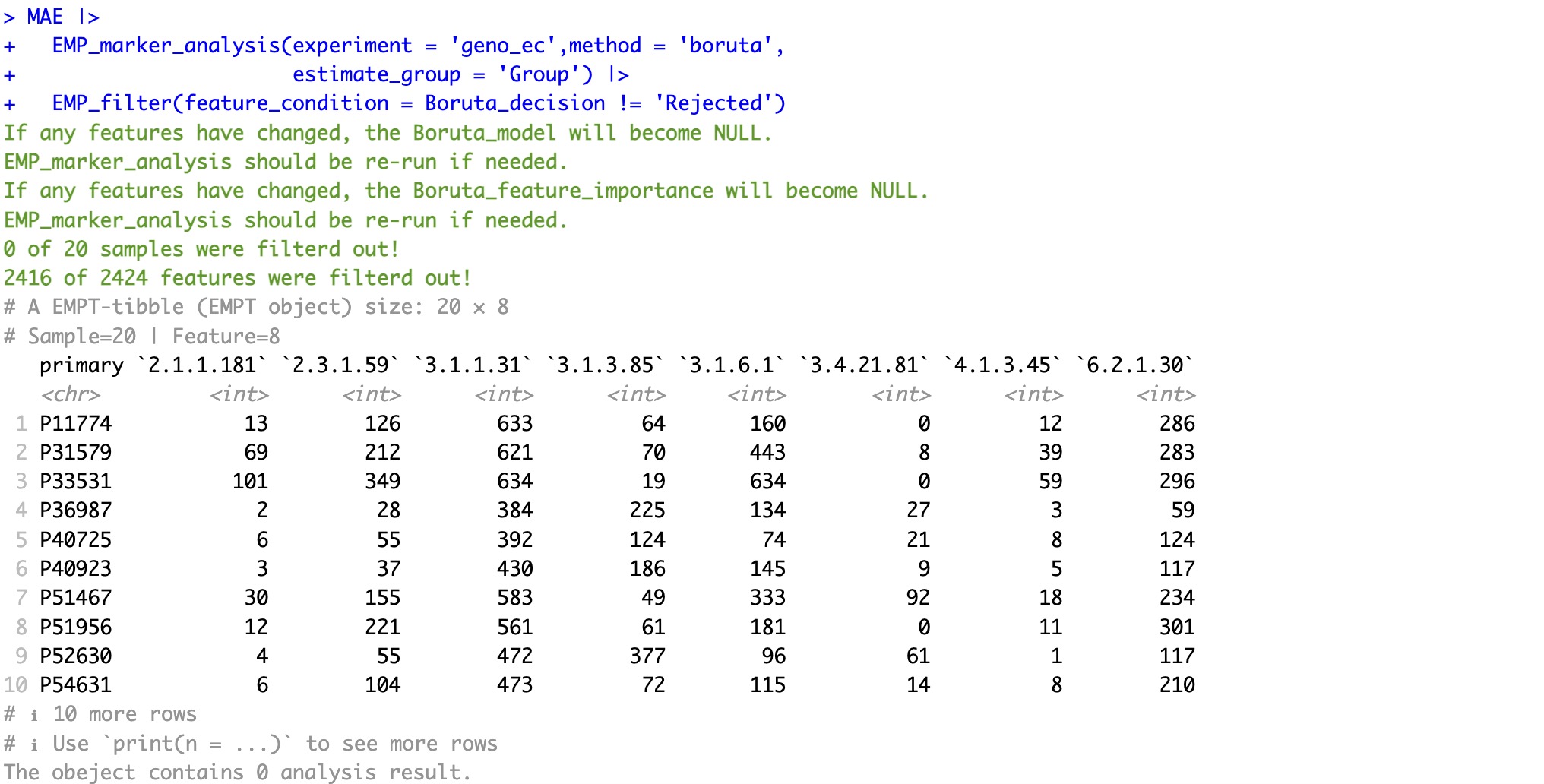
The EMP_filter module can be used to quickly filter the required features, making it convenient for downstream analysis.
The module
EMP_filter is applicable to screen for all models。
MAE |>
EMP_marker_analysis(experiment = 'geno_ec',method = 'boruta',
estimate_group = 'Group') |>
EMP_filter(feature_condition = Boruta_decision != 'Rejected')
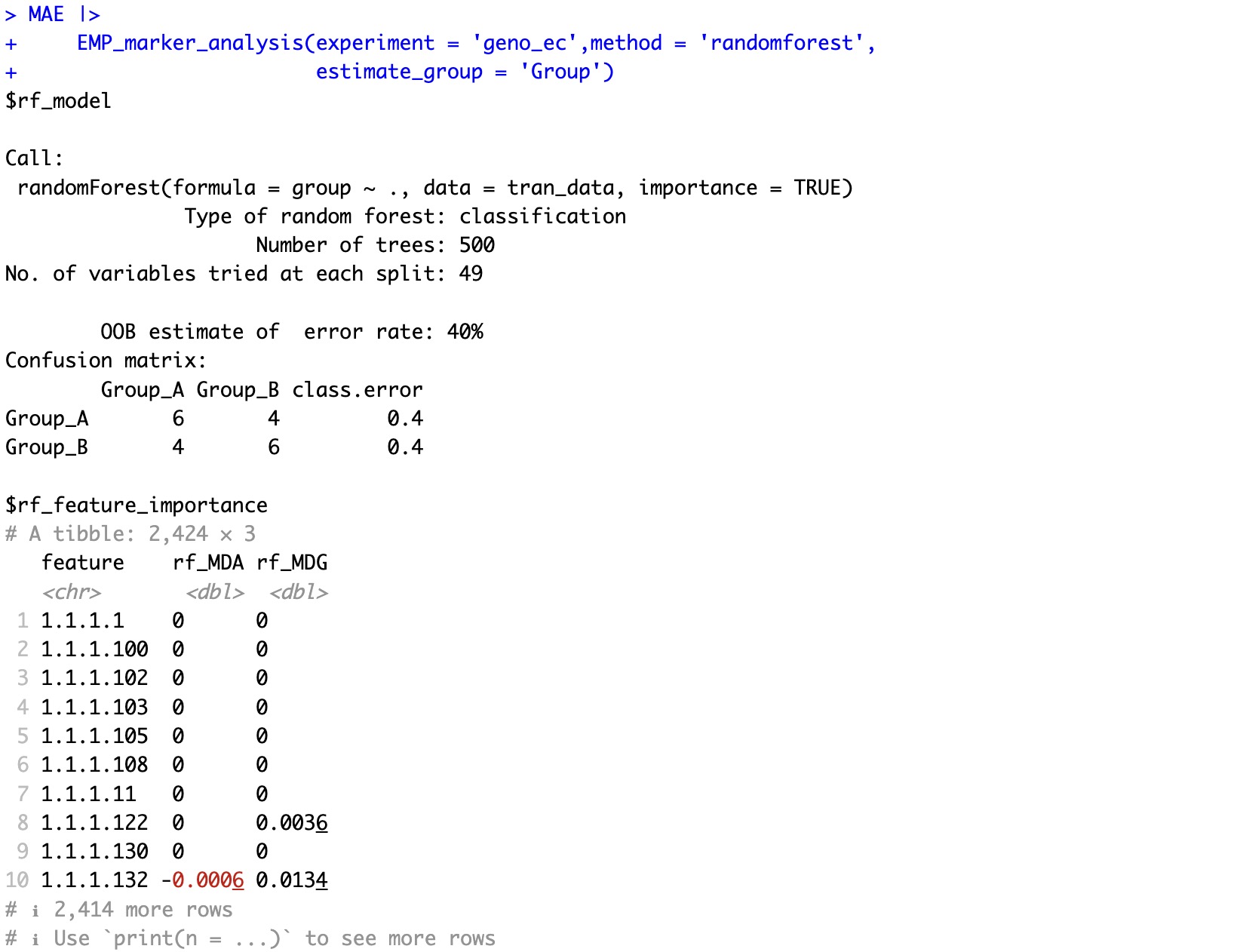
6.7.2 Estimate the importance of features based on the Random Forest Algorithm
🏷️Example1:Assessing feature importance based on random forest classification.
MAE |>
EMP_marker_analysis(experiment = 'geno_ec',method = 'randomforest',
estimate_group = 'Group')
🏷️Example2:Assessing feature importance based on random forest regression.
MAE |>
EMP_marker_analysis(experiment = 'geno_ec',method = 'randomforest',
estimate_group = 'BMI')

6.7.3 Estimate the importance of features based on the xgboost algorithm
① When using the xgboost algorithm, the model must be specified through the parameter
objective. If the value specified by the parameter estimate_group is a binary classification variable, the parameter should be set as objective = 'binary:logistic'; if the value specified by estimate_group is a multi-class classification variable, it should be set as objective = 'multi:softmax'; if the value specified by estimate_group is a continuous variable, the parameter should be set as objective = 'reg:squarederror'. More details are available in the xgboost package.② For classification models, the parameter
xgboost_run='classify' must be specified.③ For regression models, the parameter
xgboost_run='regress' must be specified.④ An incorrect specification of the parameter
xgboost_run will result in incorrect outcomes.
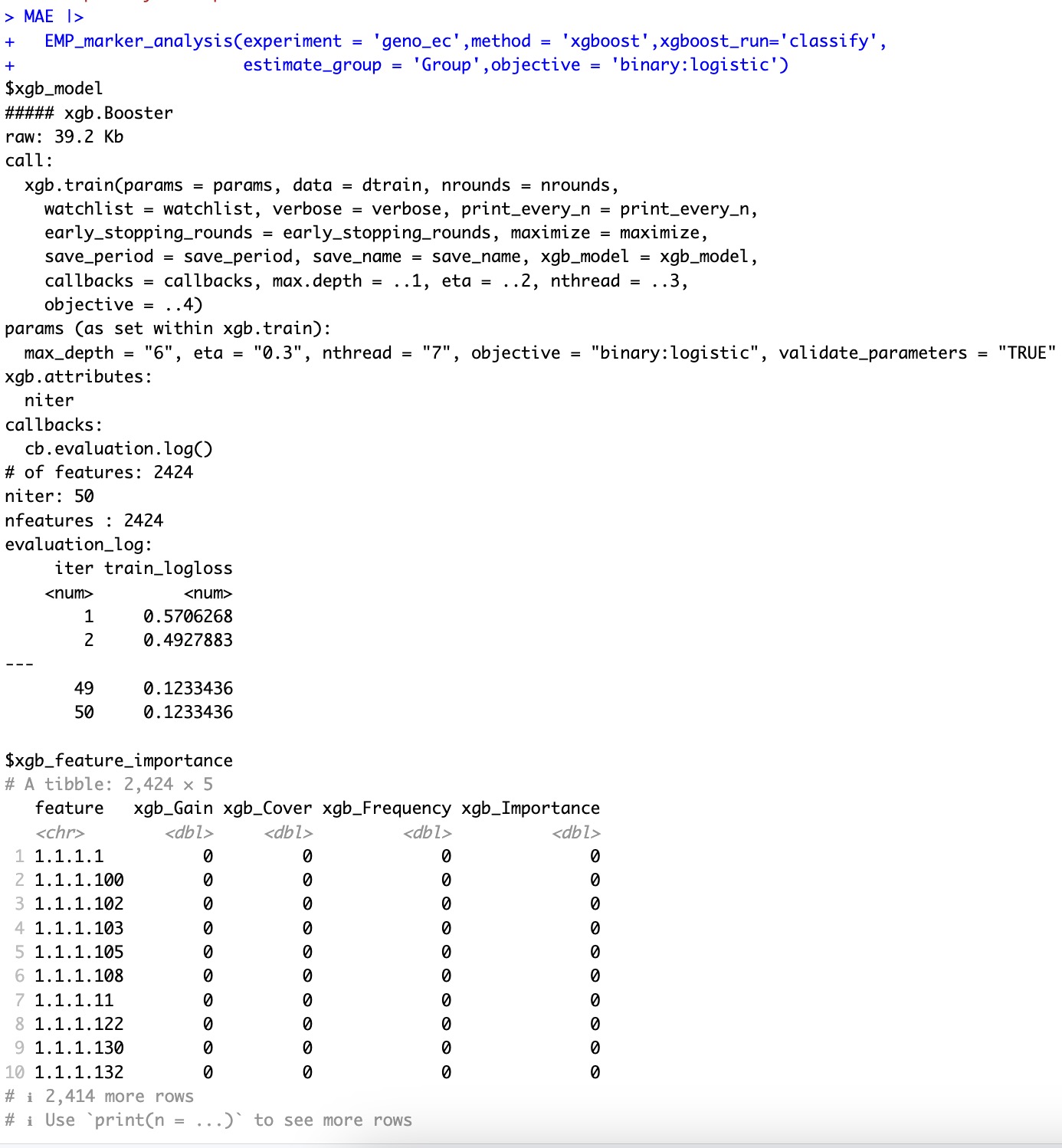
🏷️Example1:基于xgboost分类算法进行特征重要性评估。
MAE |>
EMP_marker_analysis(experiment = 'geno_ec',method = 'xgboost',xgboost_run='classify',
estimate_group = 'Group',objective = 'binary:logistic')
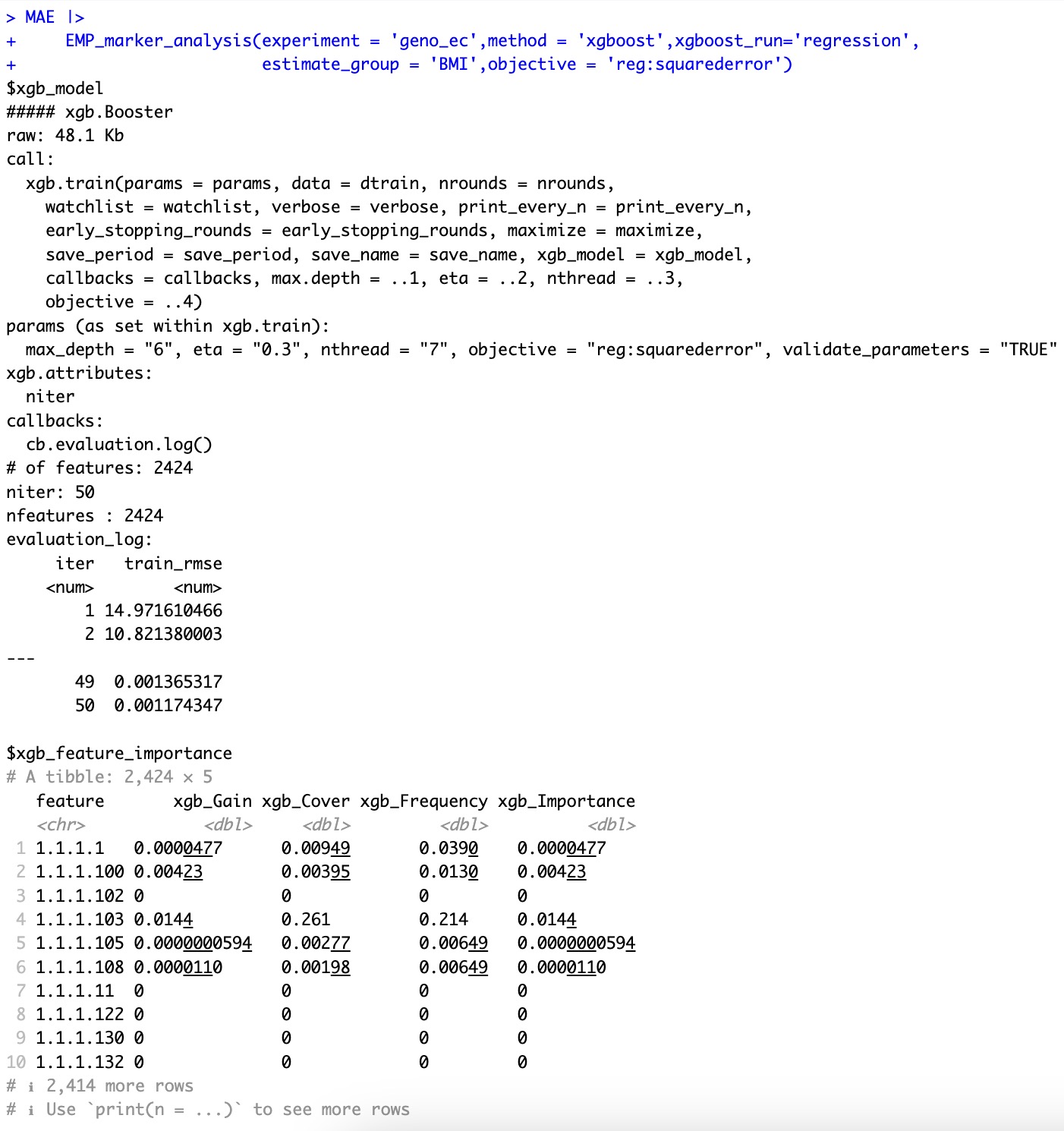
🏷️Example2:基于xgboost回归算法进行特征的重要性评估。
MAE |>
EMP_marker_analysis(experiment = 'geno_ec',method = 'xgboost',xgboost_run='regression',
estimate_group = 'BMI',objective = 'reg:squarederror')
6.7.4 基于Lasso算法估计特征的重要性
🏷️Example:
MAE |>
EMP_marker_analysis(experiment = 'geno_ec',method = 'lasso',
estimate_group = 'BMI')
